Translational Medicine
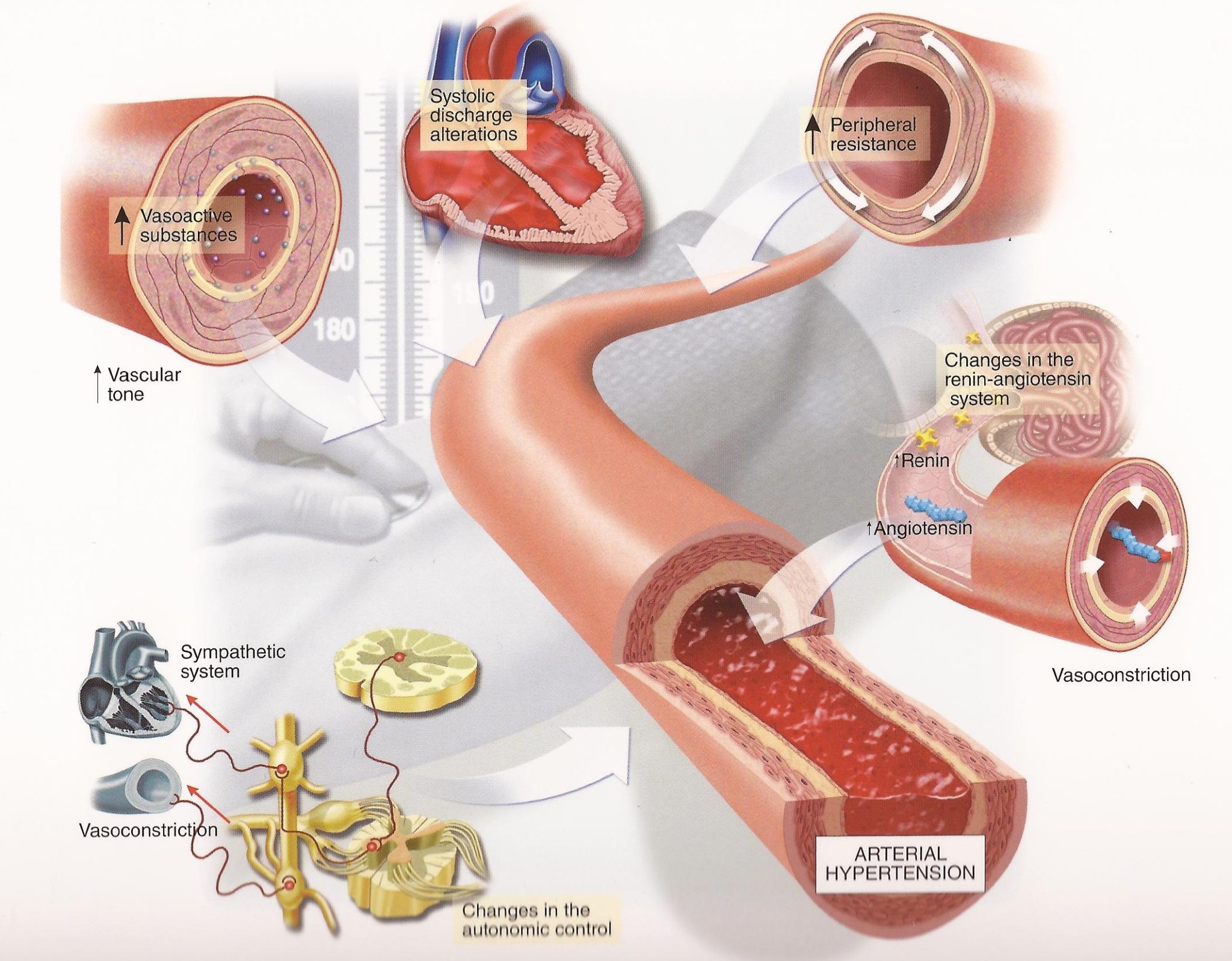
Hypertension, or high blood pressure, is a vascular disease that effects over 116 million Americans (46% of the adult population) resulting in half a million deaths and $131 billion in healthcare costs annually. Considerable research has focused on hypertension and developing pharmacological treatments to lower blood pressure. Yet, we still lack an adequate understanding of how the vascular 3D microenvironment drives disease through the complex interdependence of cell generated forces, pulsatile flow, extra cellular matrix (ECM) production, and molecular signaling. We are interested in how high blood pressure alters the structure and function of small diameter blood vessels.
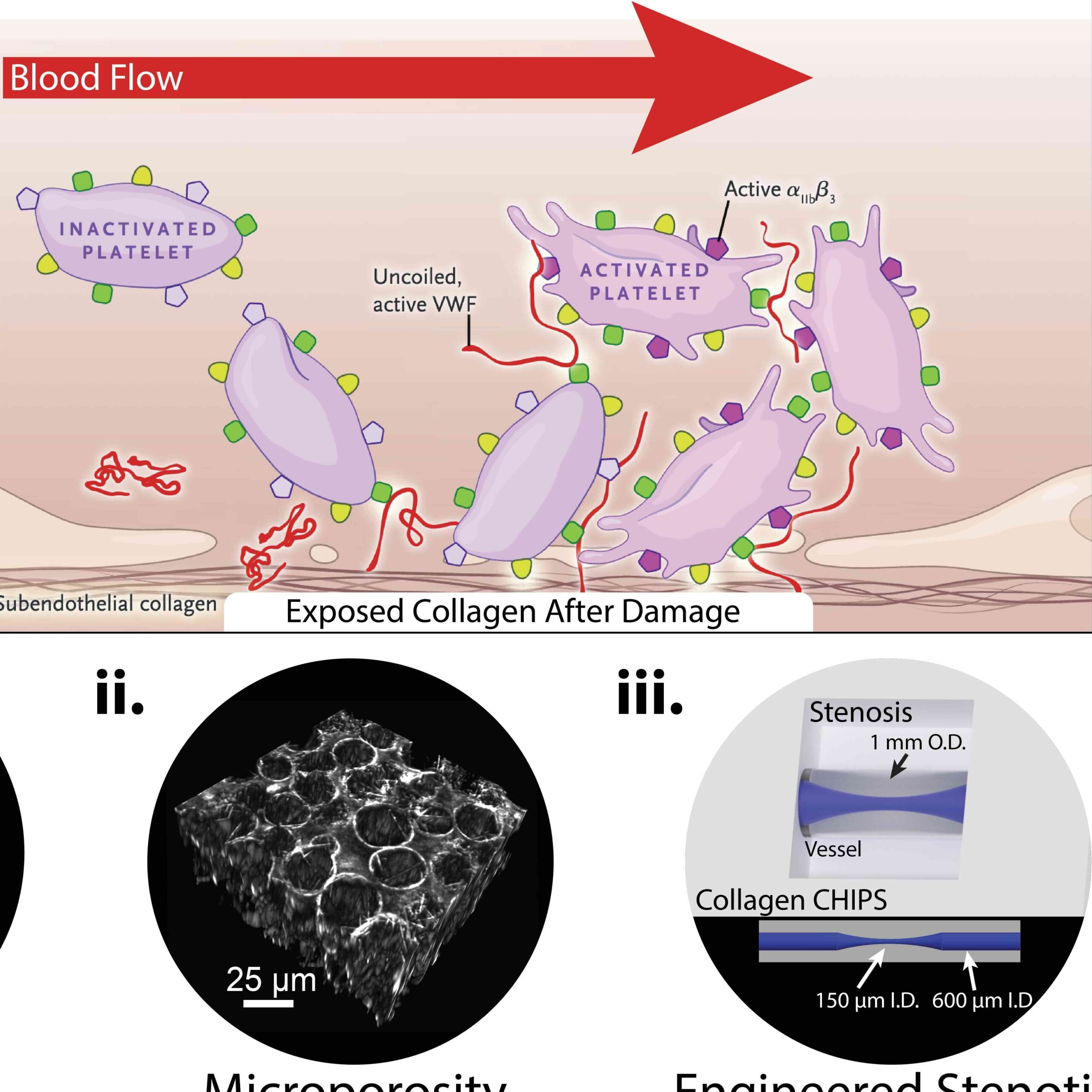
We are also starting a new line of investigation into rare clotting disorders such as Von Willebrand’s Disease and how engineered systems can be leverage for diagnostic purposes and the study of patient-specific genetic mutations. Specifically, we are 3D bioprinting collagen scaffolds to act as a platform for bleeding assessment, von Willebrand factor binding, and platelet activation.

Acellular Vessels
Studying VWF Disorders
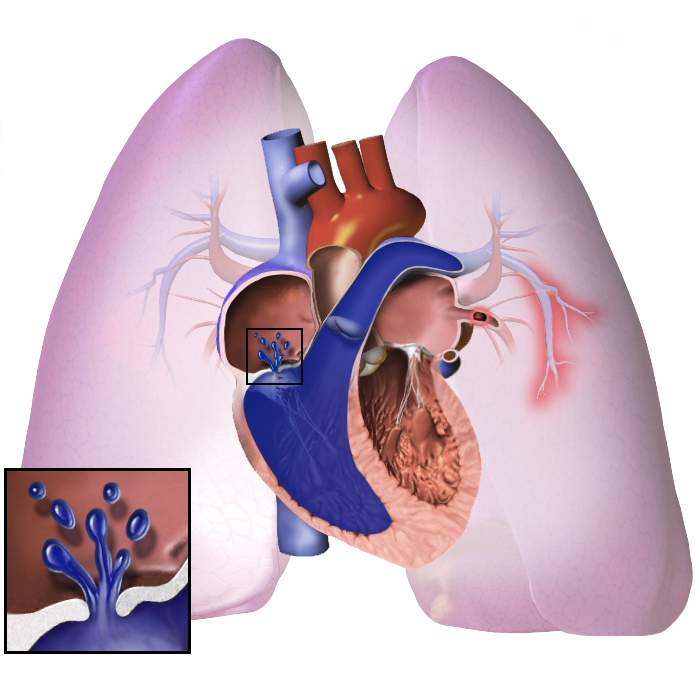
Pulmonary Hypertension
Tissue Engineering and FRESH 3D Bioprinting
The key innovation in Freeform Reversible Embedding of Suspended Hydrogels (FRESH) 3D bioprinting is the extrusion and embedding of an ECM hydrogel within a support bath that holds the extruded gel in place during printing.
In a recent publication in Science we showcased FRESH by 3D printing collagen-I into functional components of the human heart. Multiple bioinks can be printed including alginate, fibrin, collagen-I, methacrylated hyaluronic acid, decellularized ECM, and Matrigel demonstrating our ability to utilize a range of bioinks to tune scaffold composition.
Recently, we created a collagen-based 3D bioprinted vascular microfluidic platform using FRESH bioprinting (CHIPS) as well as the development of a custom perfusion bioreactor for long-term perfusion culture (VAPOR). Together, these systems establish the foundation for design and fabrication to accurately and reproducibly bioprint any 3D fluidic channel design within a collagen-scaffold. At the University of Pittsburgh in the Departments of Bioengineering and Vascular Medicine Institute we collaborate with many investigators from the School of Medicine and Carnegie Mellon University to incorporate physiological readouts, bioelectronics, and real-time fluorescence imaging into our platforms for organ-on-chip systems to study human vascular, kidney, and pancreatic disease.
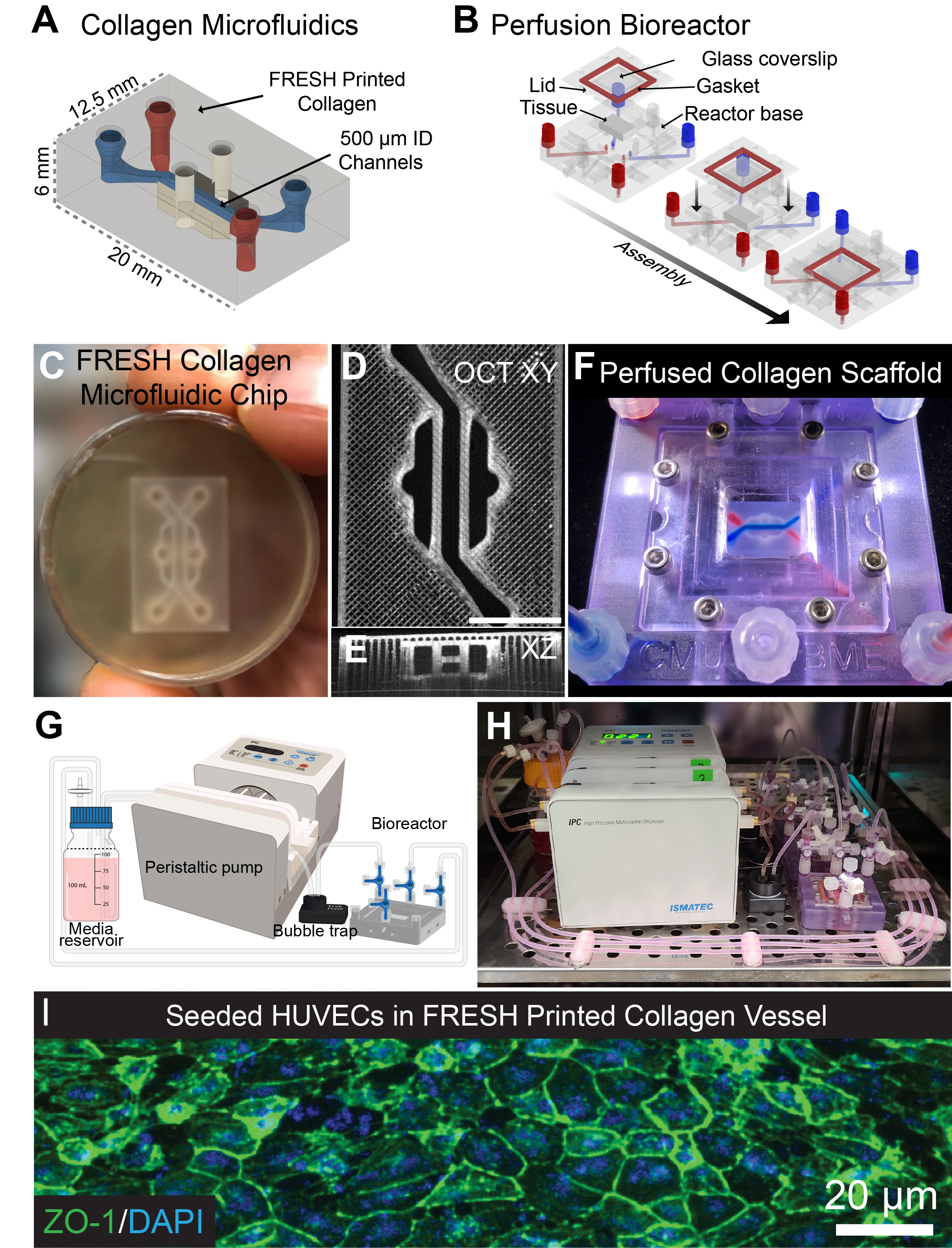
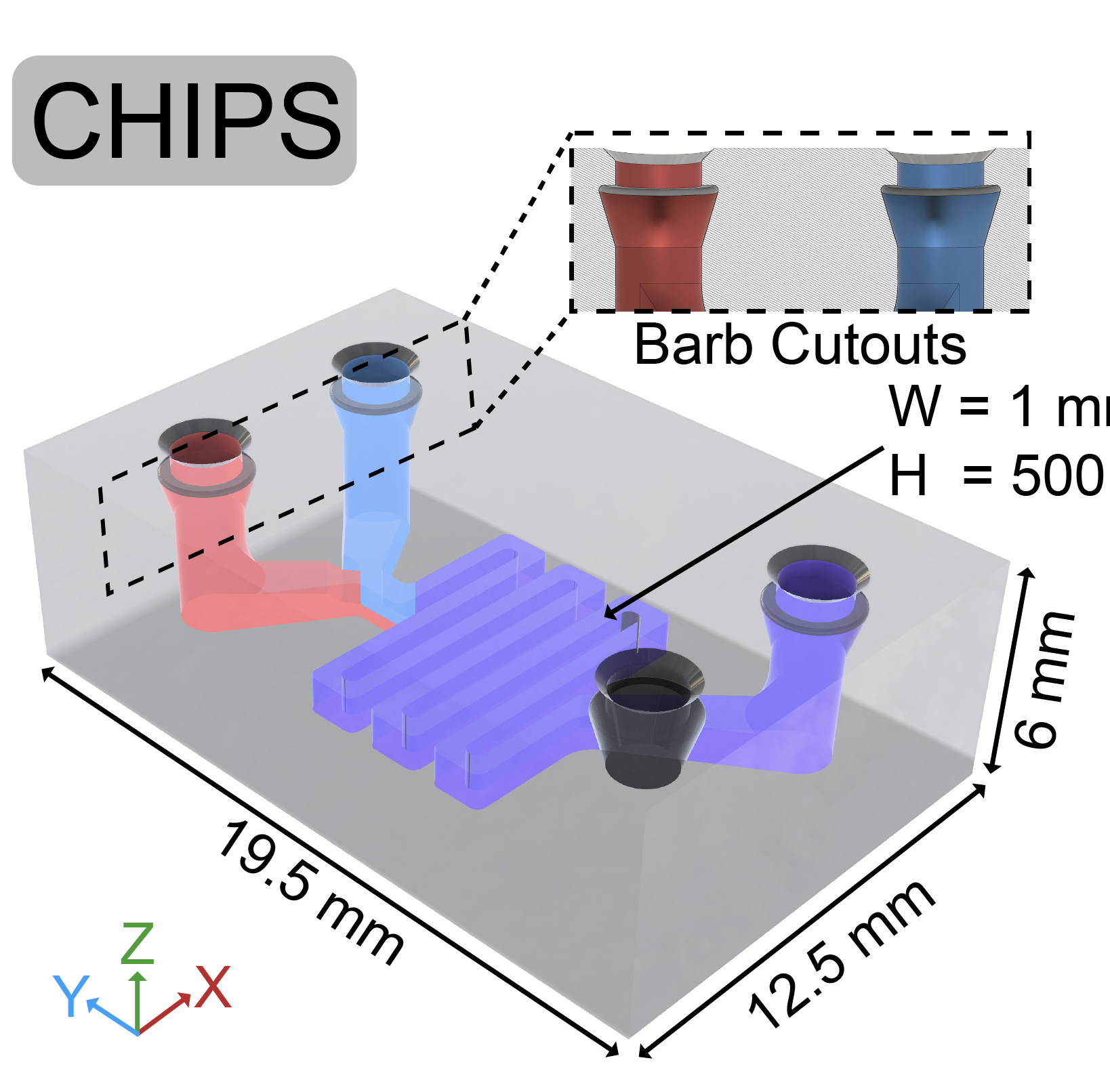
CHIPS
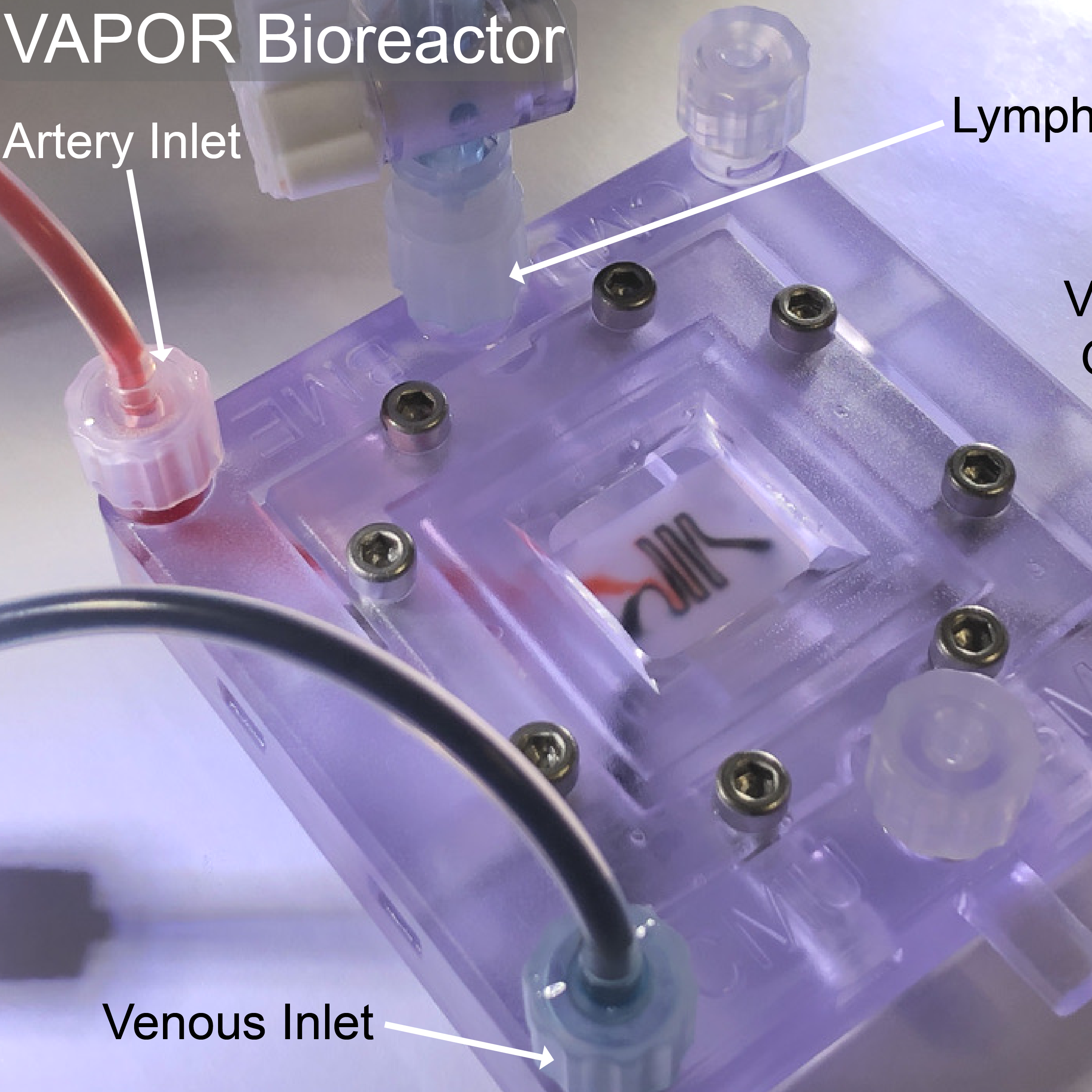
VAPOR
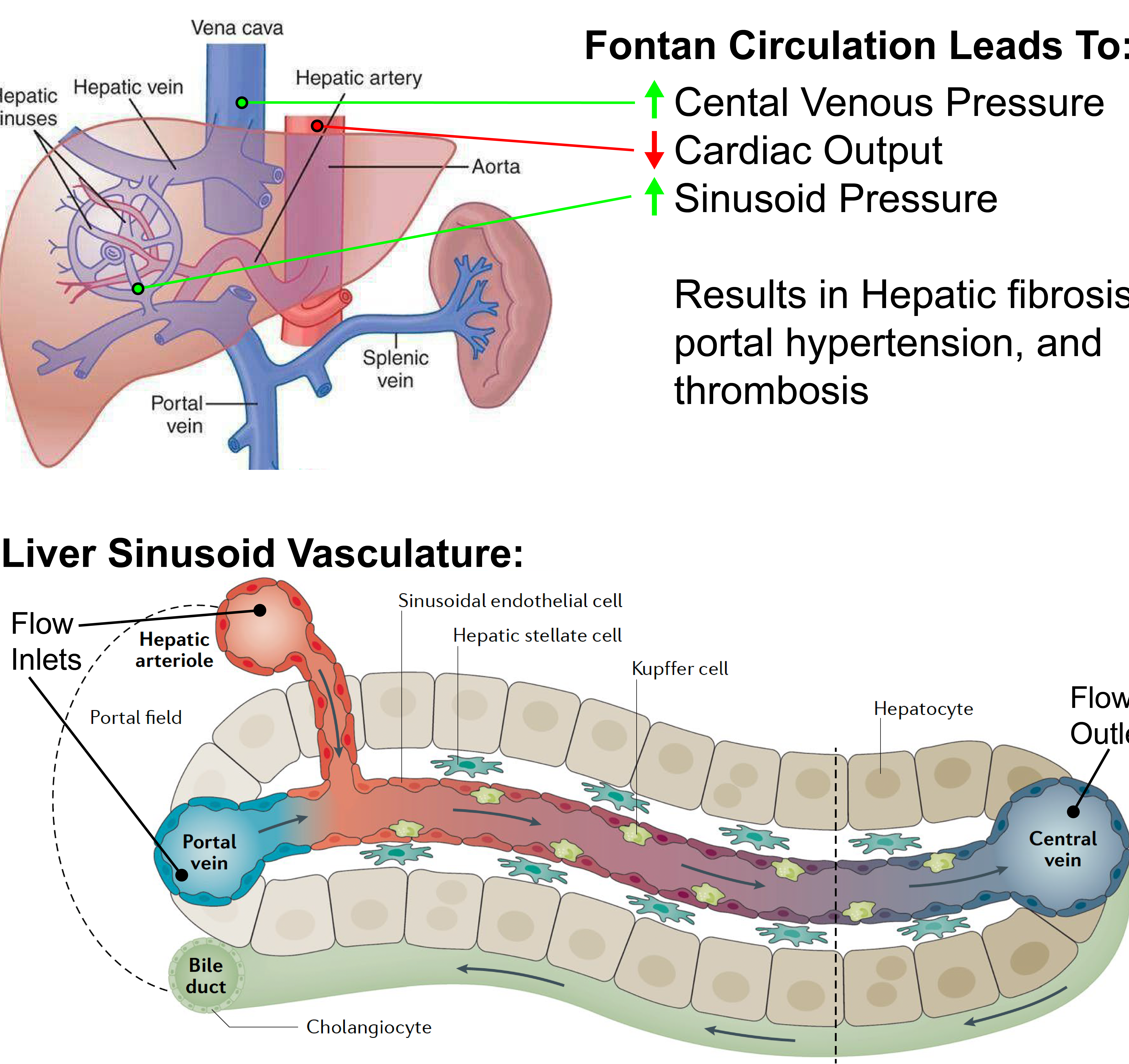
Hepatic Thrombosis
Cell Biology
One of our ongoing research interests lies in utilization of quantitative analysis of live cell fluorescence imaging to investigate basic cell biology and protein trafficking. Dr. Shiwarski is a huge proponent of the phrase “seeing is believing”, and is confident that if we can help aspiring students to see biological processes in action and quantify their results, it will ignite a passion in them scientific discovery.
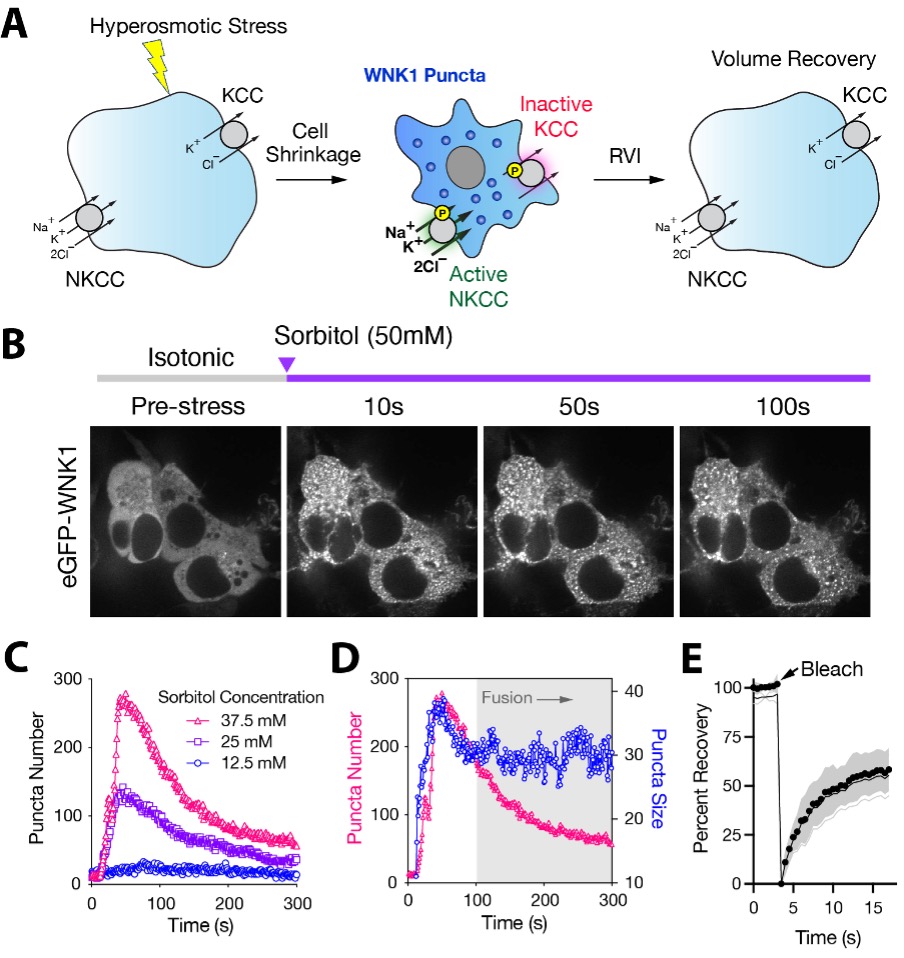
Recent work in collaboration with the Subramanya and Boyd-Shiwarski Labs at U. Pitt. has focused on leveraging live cell fluorescent imaging and quantitative analysis to study protein phase transitions of a kinase called WNK1. Using fluorescence recovery after photobleaching (FRAP), fluorescence lifetime microscopy (FLIM), and live cell particle tracking, we can measure the kinetics and mobility of WNK1 phase transitions and show that this phenomenon is a conserved and required mechanism the cell uses to regulate the activity of NKCC to achieve osmotic homeostasis.
Additionally, our group continues to utilize image quantification and analysis of fluorescent biosensors to ask novel biological questions about 3D signal propagation and developmental morphogenesis.
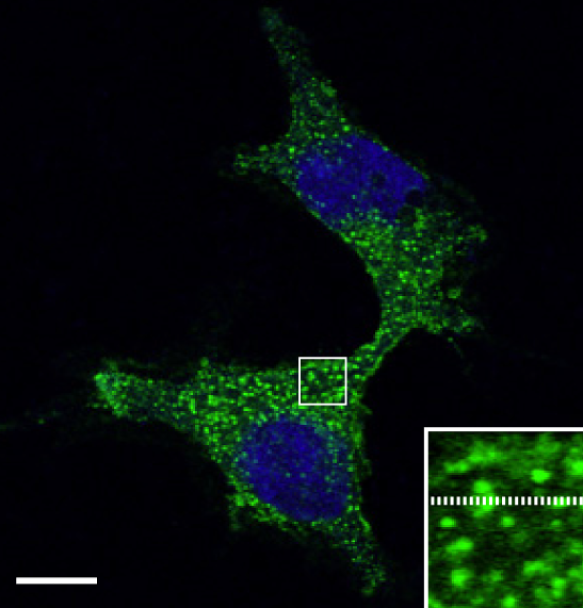
WNK1
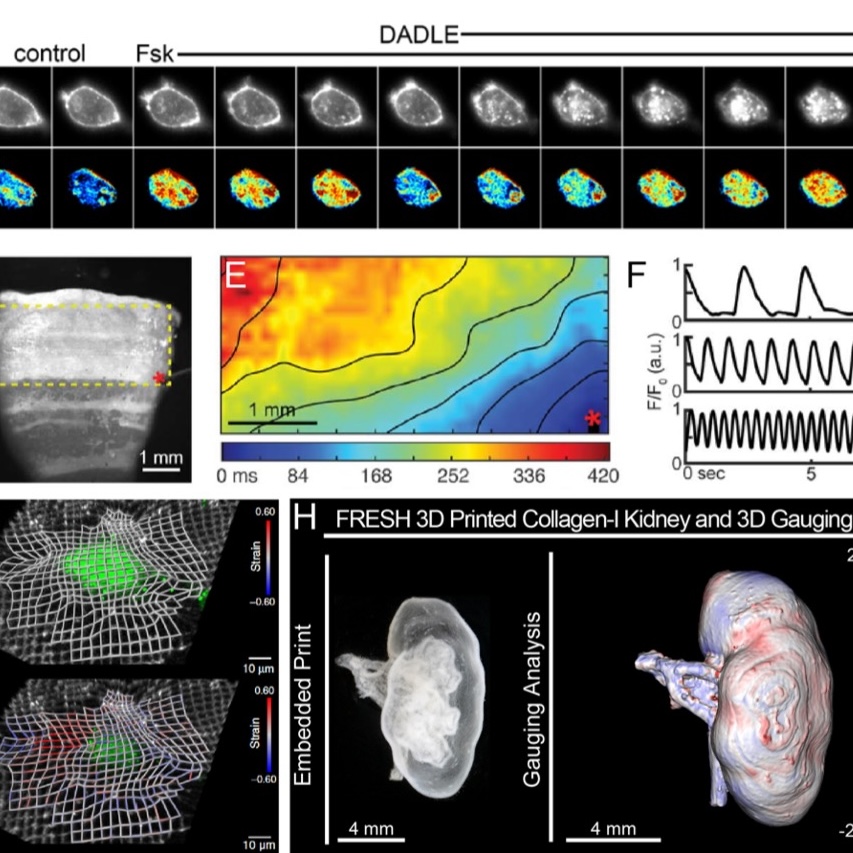
Biosensor Development
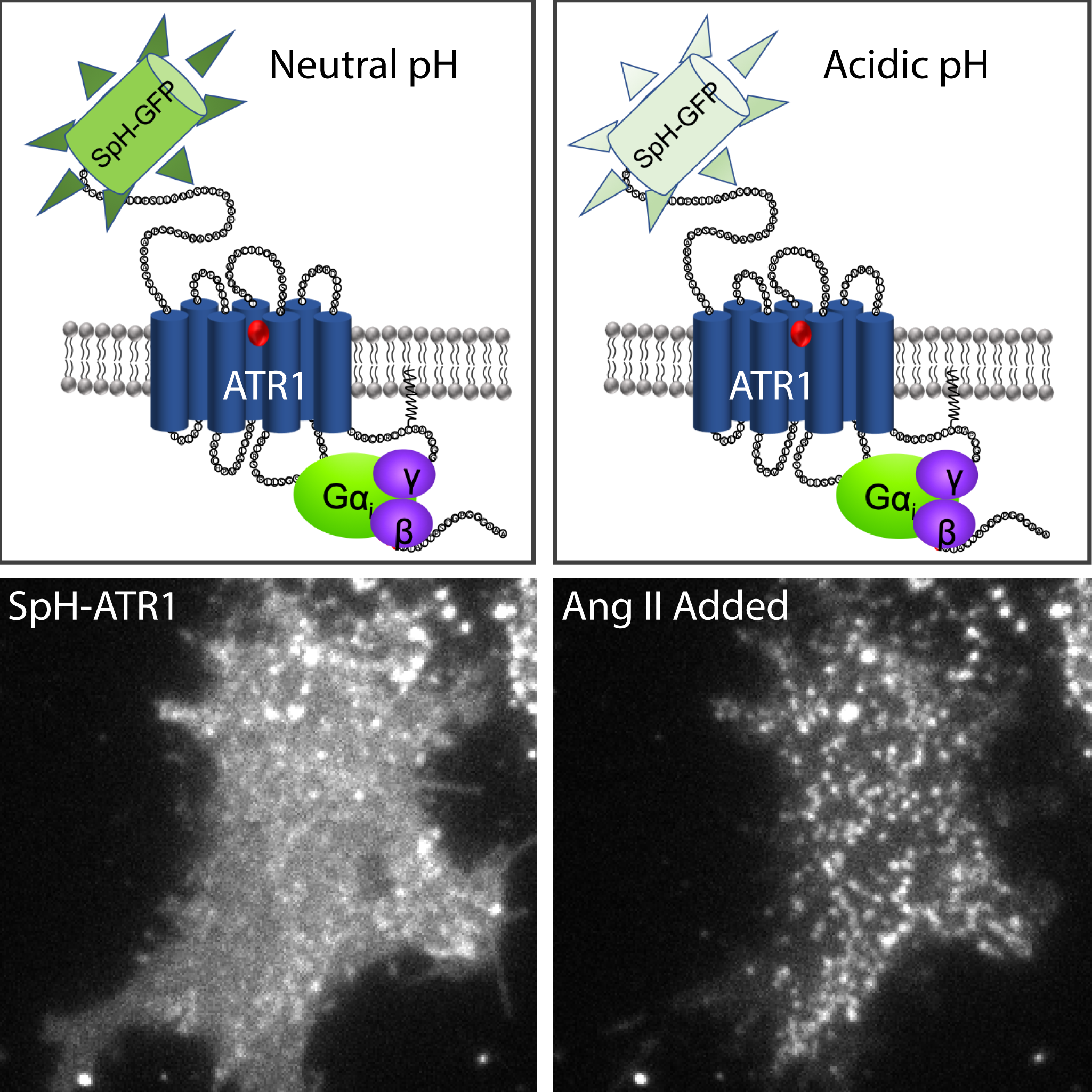
GPCR Trafficking
Open-Source Hardware
Overall, open-source hardware fosters a more inclusive and innovative approach to technology development, benefiting both individuals and the broader community. We have built bi-axial stretchers, 3D bioprinters, alignment systems, heated stages, myography devices, syringe pumps, perfusion systems, bioreactors, with many more to come.
Open-source hardware encourages collaboration and collective problem-solving. Researchers can contribute improvements, fix bugs, and innovate upon existing designs, leading to more rapid advancements and a broader range of solutions.
Open-source hardware can reduce costs for both developers and users. With freely available designs, individuals and labs can build and modify hardware without needing to invest in expensive proprietary designs or licenses.
Users have the freedom to modify and customize the hardware to meet their specific needs. This is valuable in research where bespoke solutions can be tailored to unique requirements.
Open-source hardware provides a valuable resource for education and learning. Students and hobbyists can study, build, and experiment with real-world designs, gaining hands-on experience and a deeper understanding of hardware engineering. We host annual workshops on open-source bioengineering and bioprinting. See Outreach & Education Page
A vibrant community often forms around open-source hardware projects, providing support, documentation, and shared knowledge.
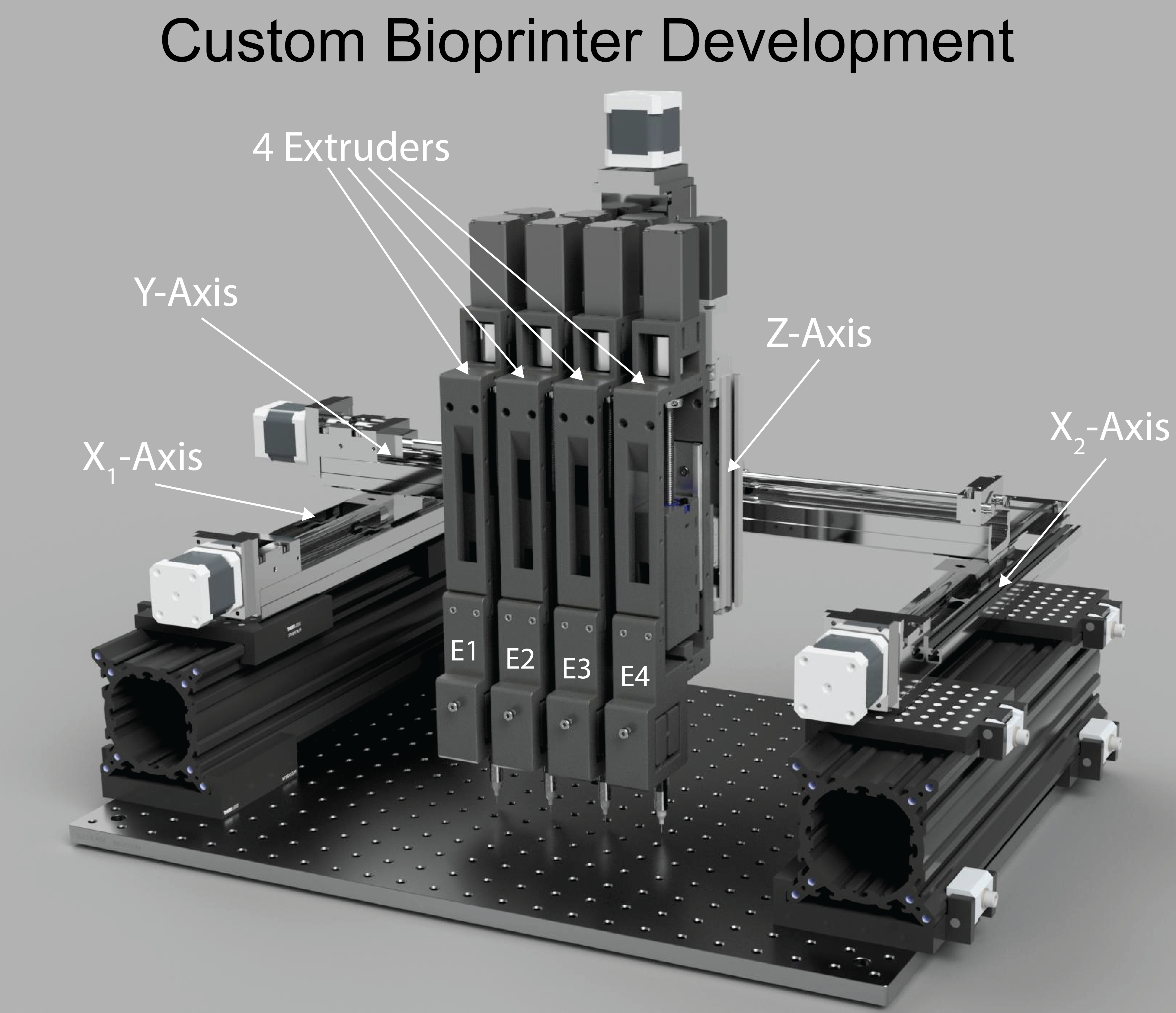

Building Bioprinters
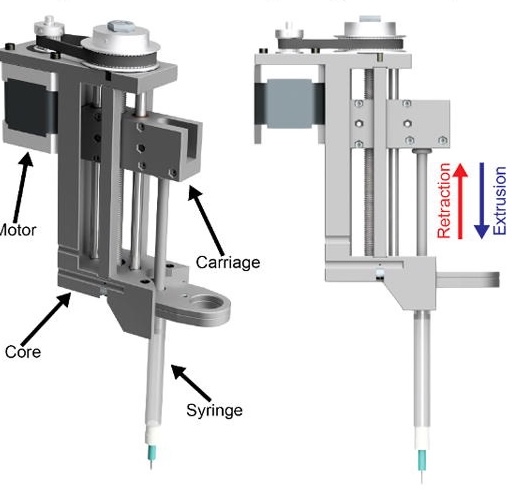
Custom Extruders


